It’s been great to see the collaborative effort from the NMS Computing Support team, our researchers, and our wider High Performance Computing (HPC) community at King’s working together to contribute valuable resources to this global Covid-19 research initiative. These additional resources will be of considerable value in contributing to the immediate drive to identify and assess the viability of novel drugs in the fight against the CoV-SARS-2 virus. Additionally, the data that will be produced as part of this initiative will underpin the various research projects that will undoubtedly result in better understanding of the molecular mechanisms responsible for viral infections.
02 April 2020
King's computer labs redeployed towards global coronavirus research
The Faculty of Natural & Mathematical Science’s (NMS) Computing Support team is taking part in a global computational effort led by Folding@home to identify potential small drug molecules in the fight against the SARS-CoV-2 virus by redeploying now idle student lab computers.

Folding@home is a distributed computing software project, running for almost 20 years, that utilises the donation of idle computing resources, such as gaming computers or consoles, to help support scientific research. Volunteers can install the programme onto their computer, which will run when the computer is not in use, for instance overnight. These collective resources are particularly valuable in running simulations of molecular interactions, which can accelerate drug discovery. Folding@home has recently started running workloads to support Covid-19 research.
To help contribute to this global effort, the NMS Computing Support team has deployed the software to a number of now idle student lab computers, and open-sourced the automation code of the machines to share with other sites looking to deploy the software to large numbers of Linux workstations.
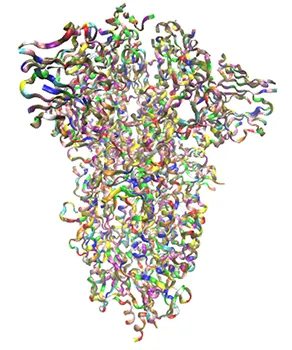
Efforts are focused at identifying small molecules to inhibit or block the Spike Protein (S Protein) of the CoV-SARS-2 virus, which acts to bind the virus to healthy cells thus leading to infection.
All protein structures that are identified via the Folding@home simulations, and all of the associated data that results from the simulations, will be made available to the greater scientific community, so that further analysis and investigation can take place.
Researchers within NMS have also been invited to contribute any idle computing resources towards the wider pool, and the Computing Support team is further exploring how the capabilities of the Folding@home software can be run on other systems.
The unsurprisingly widespread uptake of Folding@home’s recent call for more resources towards coronavirus efforts has cumulatively resulted in generating over 470 petaFLOPS of compute power, making the system the world’s fastest computer – having twice the computing power than that of the IBM Summit super computer.
Dr Chris Lorenz, Department of Physics, commented,
Find out more about how King’s is helping the fight against coronavirus.


